Help Document
About OPP_ncRBase
OPP_ncRBase(ncRNA database for oil producing plants) collected miRNAs, lncRNAs and circRNAs for oil producing plants. In additions to existing annotations for oil producing plants, we predicted many miRNAs, lncRNAs and circRNAs from high-throughput sequencing data using the following pipelines.
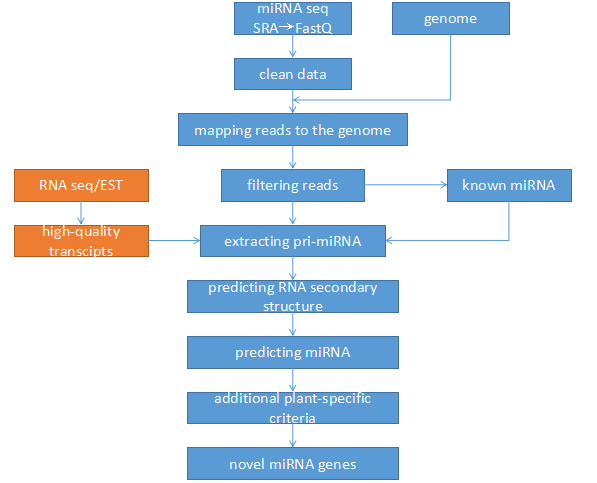
Pipeline of miRNA prediction
We use miRNA seq data from SRA(Sequence Read Archive) to prediction miRNAs for oil plants with genome. And for oil plants which haven’t genome, we use RNA seq data or EST data to prediction their miRNAs.
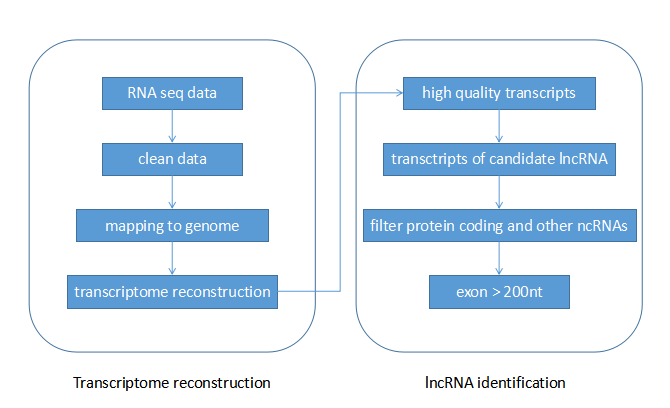
Pipeline of lncRNA prediction
We use RNA seq data from SRA to prediction lncRNAs for oil plants.
Pipeline of circRNA prediction
We used software PcircRNA_finder to prediction circRNAs for oil plants. The figure is flowchart of PcircRNA_finder for circRNA prediction. It consists of three modules.

Browse
The browsing facility allows a user to explore the data for ncRNAs by species.


Search
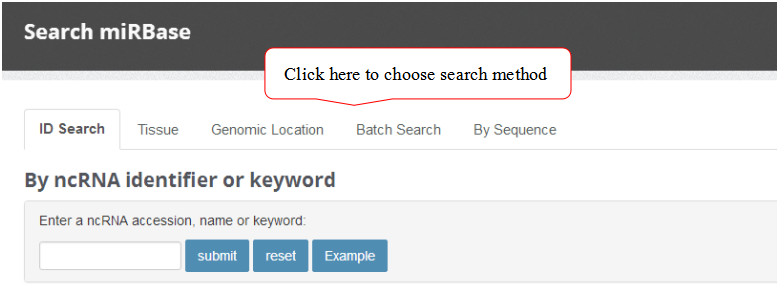
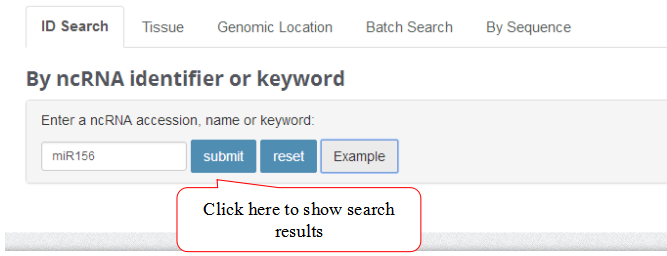
ID Search
A ID search feature allows users to search the database with keywords such as ncRNA accession, name or keywords.




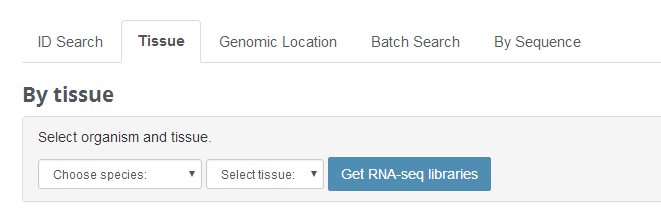
Tissue
Tissue feature allows users to search the RNA seq libraries we used to predict ncRNAs.
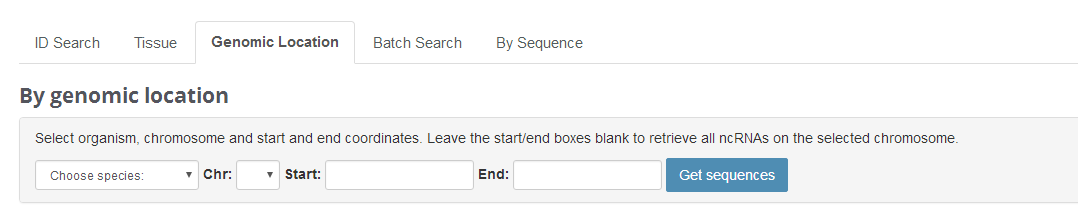
Genomic Location
Tissue feature allows users to search ncRNAs according to their genomic location.
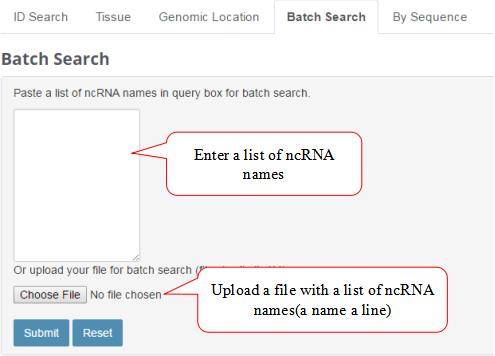
Batch Search
Batch Search feature allows users to search a lot of ncRNAs one time.
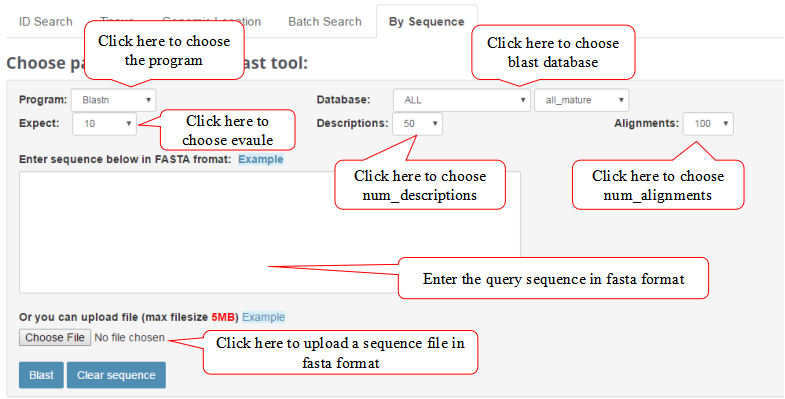
By Sequence
By Sequence feature allows users to search database by sequences.
New microRNA Prediction
New microRNA Prediction tool allows users to predict miRNAs using their own deep sequencing data.

Blast
Blast can help users to find similar sequences against their input sequences.

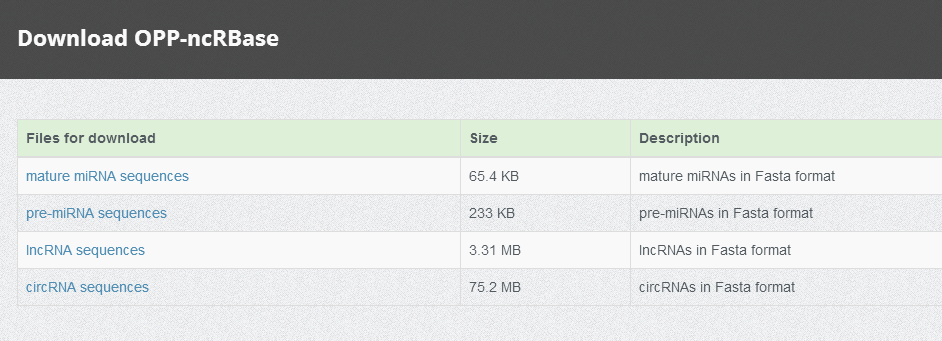
Downloads
We provide all sequences of our entries for users.